Modern high throughput DNA sequencing technology continues to revolutionize life science research. However, tens to hundreds of millions of DNA sequence records within tens of thousands of datasets aggregates into petabytes of data. HPC/HTC systems like The Open Science Grid are required to process all this data into useful data structures. OSG-GEM is a Pegasus workflow that processes DNA sequencing text files to produce a Gene Expression Matrix (GEM), which contains quantified gene expression values across tens to thousands of samples. Due to storage and memory constraints on available compute nodes, the workflow splits raw input files into small pieces to process in parallel, and merges intermediate output files. Demonstrating the portability of Pegasus workflows, OSG-GEM is configured to run on both the Open Science Grid and Jetstream. The workflow contains a configuration file that allows the user to easily specify their input dataset locations, select software options (TopHat2, HISAT2, STAR, etc.), and customize hardware requests. The output files from the workflow are formatted for input into downstream biological analysis tools, such as differential gene expression analysis and gene coexpression network construction. In addition, a statistical report is produced by the workflow that users can view to ensure the quality of their data.
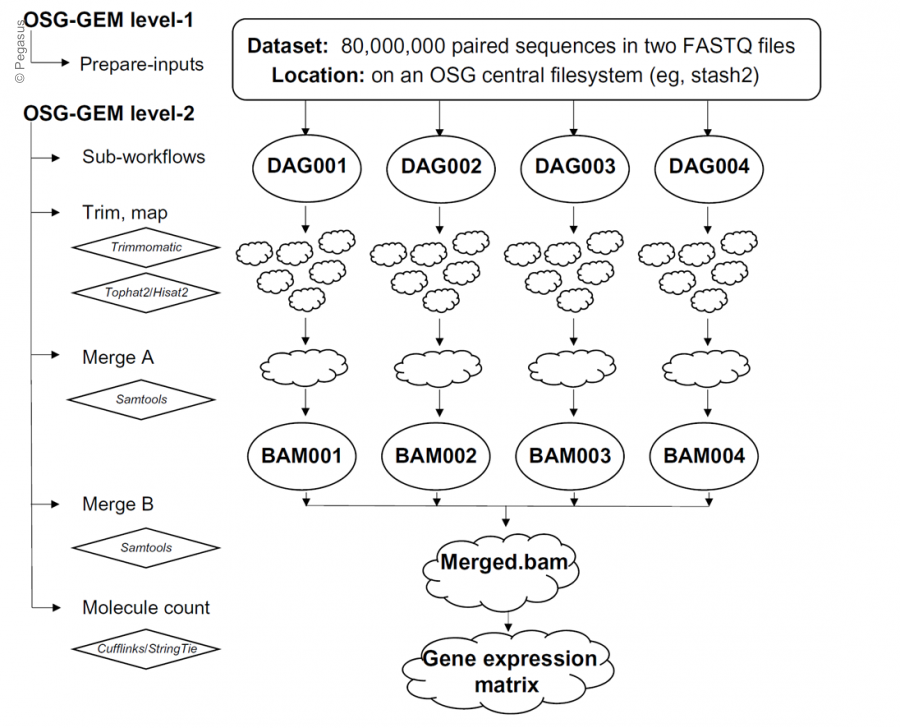
Figure copied from: William L. Poehlman, Mats Rynge, Chris Branton, D. Balamurugan and Frank A. Feltus. OSG-GEM: Gene Expression Matrix Construction Using the Open Science Grid. Bioinformatics and Biology Insights 2016:10 133–141 doi: 10.4137/BBI.S38193.
https://creativecommons.org/licenses/by/3.0/us/
Github repository: https://github.com/feltus/OSG-GEM
Scientists: William Poehlman and Alex Feltus, Clemson University