What are scientific workflows?
Scientific workflows allow users to easily express multi-step computational tasks, for example retrieve data from an instrument or a database, reformat the data, and run an analysis. A scientific workflow describes the dependencies between the tasks and in most cases the workflow is described as a directed acyclic graph (DAG), where the nodes are tasks and the edges denote the task dependencies. A defining property for a scientific workflow is that it manages data flow. The tasks in a scientific workflow can be everything from short serial tasks to very large parallel tasks (MPI for example) surrounded by a large number of small, serial tasks used for pre- and post-processing.
Pegasus overview
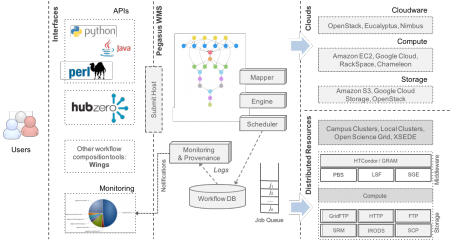
The Pegasus project encompasses a set of technologies that help workflow-based applications execute in a number of different environments including desktops, campus clusters, grids, and clouds. Pegasus bridges the scientific domain and the execution environment by automatically mapping high-level workflow descriptions onto distributed resources. It automatically locates the necessary input data and computational resources necessary for workflow execution.Pegasus enables scientists to construct workflows in abstract terms without worrying about the details of the underlying execution environment or the particulars of the low-level specifications required by the middleware (Condor, Globus, or Amazon EC2). Pegasus also bridges the current cyberinfrastructure by effectively coordinating multiple distributed resources.
Pegasus has been used in a number of scientific domains including astronomy, bioinformatics, earthquake science , gravitational wave physics, ocean science, limnology, and others. When errors occur, Pegasus tries to recover when possible by retrying tasks, by retrying the entire workflow, by providing workflow-level checkpointing, by re-mapping portions of the workflow, by trying alternative data sources for staging data, and, when all else fails, by providing a rescue workflow containing a description of only the work that remains to be done. It cleans up storage as the workflow is executed so that data-intensive workflows have enough space to execute on storage-constrained resources]. Pegasus keeps track of what has been done (provenance) including the locations of data used and produced, and which software was used with which parameters.
Pegasus has a number of features that contribute to its usability and effectiveness.
- Portability / Reuse – User created workflows can easily be run in different environments without alteration. Pegasus currently runs workflows on top of Condor, Grid infrastrucutures such as Open Science Grid and XSEDE, Amazon EC2, Google Cloud, and many campus clusters. The same workflow can run on a single system or across a heterogeneous set of resources.
- Performance – The Pegasus mapper can reorder, group, and prioritize tasks in order to increase overall workflow performance.
- Scalability – Pegasus can easily scale both the size of the workflow, and the resources that the workflow is distributed over. Pegasus runs workflows ranging from just a few computational tasks up to 1 million. The number of resources involved in executing a workflow can scale as needed without any impediments to performance.
- Provenance – By default, all jobs in Pegasus are launched using the Kickstart wrapper that captures runtime provenance of the job and helps in debugging. Provenance data is collected in a database, and the data can be queried with tools such as pegasus-statistics, pegasus-plots, or directly using SQL.
- Data Management – Pegasus handles replica selection, data transfers and output registrations in data catalogs. These tasks are added to a workflow as auxilliary jobs by the Pegasus planner.
- Reliability – Jobs and data transfers are automatically retried in case of failures. Debugging tools such as pegasus-analyzer help the user to debug the workflow in case of non-recoverable failures.
- Error Recovery – When errors occur, Pegasus tries to recover when possible by retrying tasks, by retrying the entire workflow, by providing workflow-level checkpointing, by re-mapping portions of the workflow, by trying alternative data sources for staging data, and, when all else fails, by providing a rescue workflow containing a description of only the work that remains to be done. It cleans up storage as the workflow is executed so that data-intensive workflows have enough space to execute on storage-constrained resources. Pegasus keeps track of what has been done (provenance) including the locations of data used and produced, and which software was used with which parameters.
Funding
Pegasus is funded by The National Science Foundation under OAC SI2-SSI program, grant #1664162. Previously, NSF has funded Pegasus under OCI SDCI program grant #0722019 and OCI SI2-SSI program grant #1148515.
Featured Publications
47,167 views