In 2022, USC New Cryogenic Electron Microscopy Facility officially opened for business. The facility houses two state-of-the-art electron microscopes capable of imaging molecules. CryoEM mainly focuses on structural biology and centers on studying the shapes of biological components, such as proteins and ribosomes, and how their shapes change as they perform their tasks within the cell. The field is important to understanding how molecules function within cells and to developing new therapies. An early focus of the facility, single-particle Cryo-EM allows structural biologists to visualize molecules and how they change when other molecules, such as new drugs, are present.
Unlike traditional microscopes where a researcher looks into the telescope and is able to look at the magnified samples, electron microscopes generate large amounts of data (order of Terabytes) that then needs to be processed and visualized for a researcher to inspect. The facility housing the electron microscope does not have the means to process the data generated. The microscope and its control PCs are in a dedicated facility. In order to make these microscopes widely available, the research community at USC Core Center of Excellence in Nano Imaging approached Center for Advanced Research Computing at USC (CARC) led by Byoung-Do Kim, to coordinate the development of an automated solution for transferring and processing the experimental data. CARC operates a HPC cluster that is well suited for processing these large datasets. A key aspect of electron microscopy is the fine-tuning of the data acquisitions, where preprocessing of data is done in near real time at much lower magnification allowing the researcher to decide what grids of the sample need full data acquisition. This is important in context of the amount of storage and computing resources required for processing the full dataset.
CARC leadership approached the Pegasus team at USC/ISI for assistance in developing an automated solution. Pegasus is a NSF funded project led by ISI researcher Ewa Deelman, and encompasses a set of technologies that help workflow-based applications execute in a number of different environments including desktops, campus clusters, grids, and clouds. Computer Scientists from Deelman’s team, Mats Rynge and Karan Vahi worked closely with CARC staff members Tomasz Osinski and James K. Hong, to develop a light weight image pre-processing service around the Pegasus Workflow Management System (http://pegasus.isi.edu) that kept the user interaction minimum, and offered the researcher an option to start the pre-processing right after initiating the microscope session. The users receive real-time feedback enabling them to adjust parameters during data acquisition. The service relies on Pegasus to manage the data transfers required for preprocessing and efficiently launching the tasks on CARC HPC resources optimizing for storage footprint and overall execution time of the workflows.
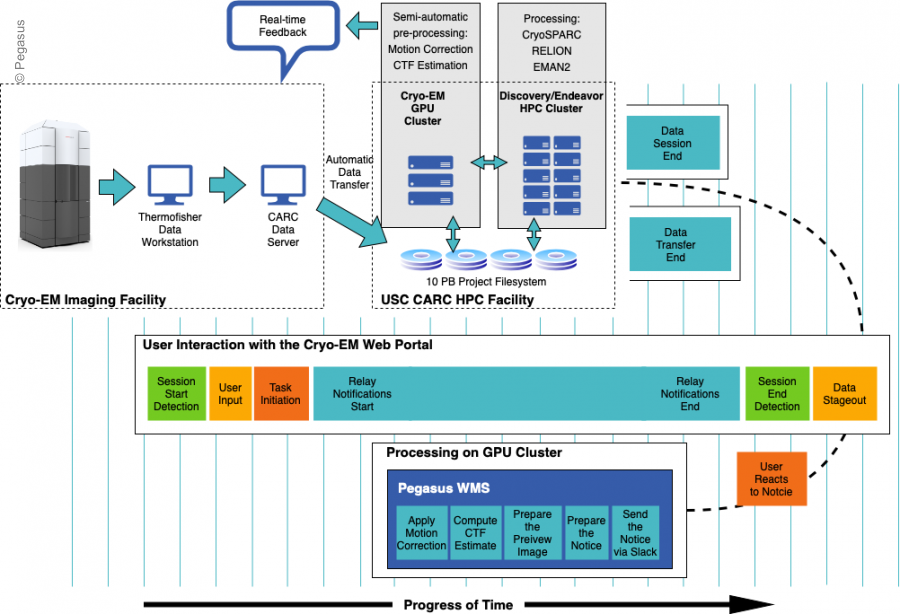
The CryoEM workflows are run on the USC HPC Cluster. For more information about the running CryoEM workflows, please see: https://www.carc.usc.edu/user-information/cryoem
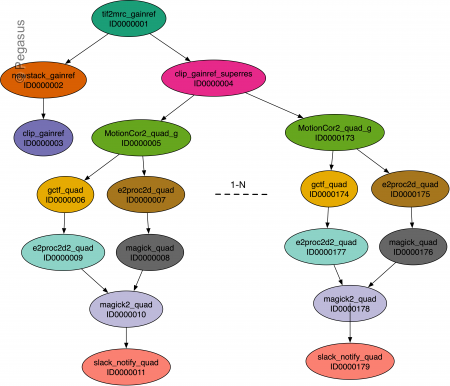
The schematic of the CryoEM processing workflow is illustrated below
Publications
Osinski, T., Rynge, M., Vahi, K., Hong, J., Chu, R., Sul, C., Deelman, E., & Kim, B.-D. (2022). An Automated Cryo-EM Computational Environment on the HPC System Using Pegasus WMS. 2022 IEEE/ACM Workflows in Support of Large-Scale Science (WORKS). (Funding Acknowledgments: )
Contacts
Tomasz Osinski : osinski@usc.edu