10. Containers
10.1. Overview
Application containers provides a solution to package software with
complex dependencies to be used during workflow execution. Starting with
Pegasus 4.8.0, Pegasus has support for application containers in the
nonsharedfs and condorio data configurations using PegasusLite.
Pegasus currently has support for the following container
technologies:
Docker
Singularity/Apptainer
Shifter
The worker package is not required to be pre-installed in images. If a matching worker package is not installed, Pegasus will try to determine which package is required and download it.
10.2. Singularity vs Apptainer
Singularity project was moved into the Linux foundation in November 2021, with an associated name change to Apptainer. To preserve backward compatibility all apptainer releases, have a symlink named singularity that points to the apptainer executable to preserve backward compatibility.
The above notwithstanding, Pegasus has been updated to prefer the apptainer executable over the singularity executable. In the case, where a job runs on a node where only an old singularity install exists, the Pegasus jobs will fallback to using the singularity executable.
In the workflow APIs; when using Pegasus you should continue to tag containers to type Singularity.
10.3. Configuring Workflows To Use Containers
Container support is provided by the Pegasus.api.transformation_catalog
Python module. Use this to generate the transformation catalog for your workflow
when containers are used.
For each container used by your workflow transformations, create a Container
and add it to your TransformationCatalog.
# Create and add containers to the TransformationCatalog.
tc = TransformationCatalog()
# A container that will be used to execute the following transformations.
tools_container = Container(
"tools-container",
Container.DOCKER,
image="docker:///ryantanaka/preprocess:latest",
arguments="--shm-size=2g",
bypass_staging=True
)
# Add the container to the TransformationCatalog
tc.add_containers(tools_container)
When this container is run, anything specified in the arguments parameter is
added as a cli argument to the resulting docker container run command that
Pegasus generates. If bypass_staging is set to True, the container will
be pulled directly onto the compute node where the job is run rather than to an
intermediate staging site first.
Then, when creating Transformations,
pass the appropriate container to the container parameter of the constructor. When is_stageable
is set to False, this means that the transformation is installed at the given
pfn inside of the container. If is_stageable is set to True, the executable will be staged into
the container and then executed there. Note that in this situation the transformation
should be hosted somewhere and made accessible via a protocol such as HTTP.
# Create and add our transformations to the TransformationCatalog.
# An executable that is installed inside of "tools_container" to be run on site "condorpool"
preprocess = Transformation(
"preprocess",
site="condorpool",
pfn="/usr/local/bin/preprocess.sh",
is_stageable=False,
container=tools_container
)
# A stageable python script that must be executed inside tools_container because
# it contains packages that we have when we developed it locally, but may not be
# installed on a compute node. Note that "pfn" is not a file URL but rather an
# http one because pegasus-transfer will need to fetch the file into the container.
process_text_2nd_pass = Transformation(
"process_text_2nd_pass.py",
site="workflow-cloud",
pfn="http://www.isi.edu/~tanaka/process_text_2nd_pass.py",
is_stageable=True,
container=tools_container
)
# Add the newly created transformations to the TransformationCatalog
tc.add_transformations(preprocess, process_text_2nd_pass)
10.3.1. Configuring Workflows To Use HTCondor Container Universe
Pegasus introduced support for containers in 4.8.0 in 2017. At that time, HTCondor did not have first class support for containers. In addition, any support for containers was tied to when running in pure HTCondor environments, and not available for example when running in grid universe (used for submitting jobs to local HPC clusters). Keeping this in mind, and also to have the most flexibility we decided to manage the staging of the application container for the job in the PegasusLite job wrapper that gets invoked when a job is launched in the worker node. This allowed us to bring container support to all the environments Pegasus supports.
While these limitations still exist, HTCondor support for containers has greatly improved in pure HTCondor environments such as PATh and OSPool; or if you are running in your local condor pool or launching condor pilots on remote resources (cloud or hpc) to overlay a condor pool.
It is important to note that container universe in HTCondor does not work when you are using HTCondor to submit jobs to another resource manager such as SLURM. For deployments, where you are submitting jobs directly to a local HPC resource, or using SSH/BOSCO to submit to a remote HPC resource, you should let Pegasus manage the container deployment on the worker nodes. The table below summarizes it.
Execution Environments |
Use Container Universe |
|---|---|
Local HTCondor Pool on a desktop. |
YES |
HTCondor pool provisioned by launching
pilots on HPC or Cloud resources.
|
YES |
HTCondor managed distributed computing
infrastructure such as OSG/OSPool/PATh.
|
YES |
Local HPC Cluster. |
NO |
Remote HPC Cluster with direct job
submission using SSH/BOSCO.
|
NO |
Cloud resources such as AWS Batch. |
NO |
Keeping this in mind, Pegasus now has support for container universe in Pegasus whereby we delegate the container management (invoking the container, launching the user job, stopping the container) to HTCondor for those environments. In order to enable this, you would need to add a condor profile with key universe and value set to “container” for your execution site in the Site Catalog.
A sample YAML snippet is included below
- name: condorpool
arch: x86_64
os.type: linux
directories: []
profiles:
condor: {universe: container}
pegasus: {style: condor, clusters.num: 1}
Note
We only support Apptainer/Singularity containers to be launched in the container universe. When enabled, Pegasus will stage-in the container as part of the data stage-in nodes in the executable workflow, and place them in the user submit directory. The container then gets transferred to the worker node where the user job is launched using in-built HTCondor file transfers.
Symlinking of Data
In case you are symlinking of data or using the shared filesystem for data staging and want those directories to be mounted in the container, you need to update the EP (Execution Point) condor configuration to specify the variable SINGULARITY_BIND_EXPR . More details can be found in the HTCondor documentation here .
10.4. Containers on OSG / PATh
If you are running worklfows on OSG or PATh, we now recommend you to use container universe to let HTCondor manage the deployment of the containers your job requires. More details can be found in the previous section about use of container universe.
10.5. Containers on HPC Clusters
Support for containers in Pegasus is available as long as the jobs are launched via PegasusLite i.e. the data configuration is either nonsharedfs or condorio .
However, when running on a HPC cluster a user may prefer to use the sharedfs data configuration that does not wrap a job with PegasusLite. A side effect of this configuration is that you can also not run jobs using a container in the sharedfs mode.
For this case, we recommend using a hybrid data staging configuration whereby you run the workflows in the nonsharedfs mode, but for your compute site you use the shared filesystem as the data staging location. Details on this configuration can be found here.
10.6. Container Execution Model
Containerized applications in your workflow are launched as part of PegasusLite jobs when starting on a remote worker node. The container execution model is as follows:
Sets up a directory to run a user job in
Pulls the container image to that directory
Optionally, loads the container from the container image file and sets up the user to run as in the container (only applicable for Docker containers)
Mounts the job directory into the container as
/scratchfor Docker containers and as/srvfor Apptainer/Singularity containersIf the data transfers for the job are set to happen on the host os (which is the default), pulls in all the relevant input data and executables required by the job in a directory on the HOST OS, that gets mounted in the container.
Container will run a job specific script created by PegasusLite that does the following:
Figures the appropriate Pegasus worker to use in the container if not already installed.
Sets up the job environment to use including transfer and setup of any credentials transferred as part of PegasusLite.
If the data transfers for the job are set to happen inside the container; pulls in all the relevant input data and executables required by the job.
Launches the user application using pegasus-kickstart.
If the data transfers for the job are set to happen inside the container; stages out the output data to the staging site.
Optionally, shuts down the container (only applicable for Docker containers)
Ships out the output data to the staging site
Cleans up the directory on the worker node
Note
Starting in Pegasus 5.1.0 release, the default for the container data transfer model has been changed back to the default that was for 5.0.0 release. The data transfers for the job occurring outside the container in the PegasusLite wrapper, instead happening inside the container, when the user job starts in the container. However, this behavior can be controlled by setting the property pegasus.transfer.container.onhost in the properties file.
10.7. Data Transfers for Jobs when running in Container
When a job is specified to run in an application container such as Docker or Singularity, Pegasus has two options in PegasusLite on how data transfers for the job occur. The transfers can happen either
on the HOST OS before the container in which the job has to execute is invoked OR
inside the application container, before the user code is invoked.
You can control the behavior by setting the Boolean property pegasus.transfer.container.onhost in your properties file.
10.7.1. HOST OS Container Transfers
By default, Pegasus will set up the data transfers to happen on the HOST OS in the PegasusLite wrapper before the application container is launched. Benefits of this approach is that, it does not require Python3 to be installed in the container. Python3 is required to run the pegasus data transfer tools such as pegasus-transfer.
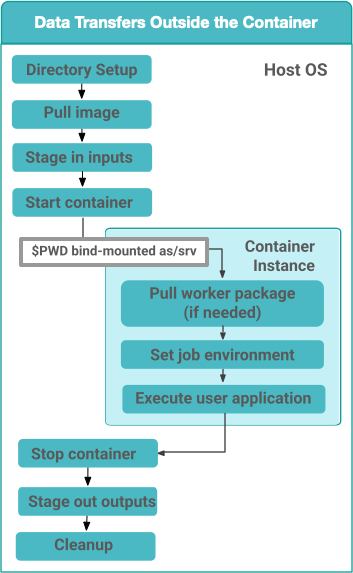
Figure 1: Data Transfers setup to be on the HOST OS
10.7.2. Data Transfers inside the Container
If the property pegasus.transfer.container.onhost is set to false, then the data transfers for the job happen inside the container before user code is invoked. In this case, the container must have Python3 available. This approach does have the benefit of you installing your own data transfer tools in the container that are not available on the HostOS and to use them for data transfers.
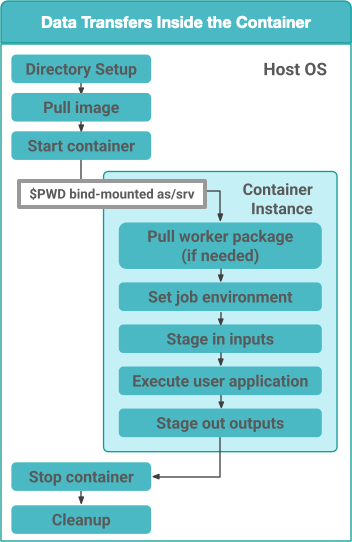
Figure 1: Data Transfers setup to be inside the Container
Since in this case, the transfers are handled from within the container, and thus container recipes require some extra attention. A Dockerfile example that prepares a container for GridFTP transfers is provided below.
In this example there are three sections.
Essential Packages
Install Globus Toolkit
Install CA Certs
From the Essential Packages, python and either curl or
wget have to be present. Install Globus Toolkit, sets up the
enviroment for GridFTP transfers. And Install CA Certs copies the grid
certificates in the container.
Note
Globus Toolkit introduced some breaking changes in August 2018 to its
authentication module, and some sites haven’t upgraded their
installations (eg. NERSC). GridFTP in order to authenticate
successfully, requires the libglobus-gssapi-gsi4 package to be pinned
to the version 13.8-1. The code snipet below contains installation
directives to handle this but they are commented out.
##########################################
#### This Container Supports GridFTP ####
##########################################
FROM ubuntu:18.04
#### Essential Packages ####
RUN apt-get update &&\
apt-get install -y software-properties-common curl wget python unzip &&\
rm -rf /var/lib/apt/lists/*
#### Install Globus Toolkit ####
RUN wget -nv http://www.globus.org/ftppub/gt6/installers/repo/globus-toolkit-repo_latest_all.deb &&\
dpkg -i globus-toolkit-repo_latest_all.deb &&\
apt-get update &&\
# apt-get install -y libglobus-gssapi-gsi4=13.8-1+gt6.bionic &&\
# apt-mark hold libglobus-gssapi-gsi4 &&\
apt-get install -y globus-data-management-client &&\
rm -f globus-toolkit-repo_latest_all.deb &&\
rm -rf /var/lib/apt/lists/*
#### Install CA Certs ####
RUN mkdir -p /etc/grid-security &&\
cd /etc/grid-security &&\
wget -nv https://download.pegasus.isi.edu/containers/certificates.tar.gz &&\
tar xzf certificates.tar.gz &&\
rm -f certificates.tar.gz
##########################################
#### Your Container Specific Commands ####
##########################################
10.8. Staging of Application Containers
Pegasus treats containers as other files in terms of data management. A container to be used for a job is tracked as an input dependency that needs to be staged if it is not already there. Similar to executables, you specify the location for your container image in the Transformation Catalog. You can specify the source URL’s for containers as the following.
URL to a container hosted on a central hub repository
Example of a docker hub URL is
docker:///rynge/montage:latest, while for singularityshub://pegasus-isi/fedora-montageURL to a container image file on a file server
Docker -Docker supports the loading of a container from a tar file. Hence, containers images can only be specified as tar files and the extension for the filename is not important.
Singularity - Singularity supports container images in various forms and relies on the extension in the filename to determine what format the file is in. Pegasus supports the following extensions for singularity container images
.img
.tar
.tar.gz
.tar.bz2
.cpio
.cpio.gz
.sif
Singularity will fail to run the container if you don’t specify the right extension when specifying the source URL for the image.
In both the cases, Pegasus will place the container image on the staging
site used for the workflow, as part of the data stage-in nodes, using
pegasus-transfer. When pulling in an image from a container hub
repository, pegasus-transfer will export the container as a tar file in
case of Docker, and as .img file in case of Singularity
10.8.1. Shifter Containers
Shifter containers are different from Docker and Singularity with respect to the fact that the containers cannot be exported to a container image file that can reside on a filesystem. Additionally, the containers are expected to be available locally on the compute sites in the local Shifter registry. Because of this, Pegasus does not do any transfer of Shifter containers. You can specify a Shifter container using the Shifter url scheme. For example, below is a transformation catalog for a namd transformation that is executed in a Shifter container.
tc = TransformationCatalog()
namd_image = Container(
"namd_image",
Container.SHIFTER,
image="shifter:///papajim/namd_image:latest"
)
tc.add_containers(namd_image)
namd_tr = Transformation(
"namd_tr",
site="cori",
pfn="/opt/NAMD_2.12_Linux-x86_64-multicore/namd2",
is_stageable=False,
container=namd_image
)\
.add_globus_profile(max_time=20)\
.add_pegasus_profile(exitcode_success_msg="End of program")
tc.add_transformations(namd_tr)
10.8.2. Symlinking and File Copy From Host OS
Since, Pegasus by default only mounts the job directory determined by PegasusLite into the application container, symlinking of input data sets works only if in the container definition in the transformation catalog user defines the directories containing the input data to be mounted in the container using the mounts key word. We recommend to keep the source and destination directories to be the same i.e. the host path is mounted in the same location in the container.
The above is also true for the case, where your input datasets are on the shared filesystem on the compute site and you want a file copy to happen, when PegasusLite job starts the container.
For example in the example below, we have input datasets accessible on
/lizard on the compute nodes, and mounting them as read-only into the
container at /lizard
centos_base = Container(
"centos-base",
Container.SINGULARITY,
image="gsiftp://bamboo.isi.edu/lfs1/bamboo-tests/data/centos7.img",
image_site="local",
mounts=["/lizard:/lizard:ro"]
).add_env(JAVA_HOME="/opt/java/1.6")
To enable symlinking for containers set the following properties
props = Properties()
# Tells Pegasus to try and create symlinks for input files
props["pegasus.transfer.links"] = "true"
# Tells Pegasus to by the staging site ( creation of stage-in jobs) as
# data is available directly on compute nodes
props["pegasus.transfer.bypass.input.staging"] = "true"
props.write()
If you don’t set pegasus.transfer.bypass.input.staging then you still can
have symlinking if:
your staging site is same as your compute site
the scratch directory specified in the site catalog is visible to the worker nodes
you mount the scratch directory in the container definition, NOT the original source directory.
Enabling symlinking of containers is useful when running large
workflows on a single cluster. Pegasus can pull the image from the
container repository once, and place it on the shared filesystem where
it can then be symlinked from when the PegasusLite jobs start on the
worker nodes of that cluster. In order to do this, you need to be
running the nonsharedfs data configuration mode with the staging site
set to be the same as the compute site.