1. Introduction
Pegasus WMS is a configurable system for mapping and executing scientific workflows over a wide range of execution environments including a laptop, a campus cluster, a Grid, or a commercial or academic cloud. Today, Pegasus runs workflows on Amazon EC2, Nimbus, Open Science Grid, and many campus clusters. One workflow can run on a single system or across a heterogeneous set of resources. Pegasus can run workflows ranging from just a few computational tasks up to 1 million.
Pegasus WMS bridges the scientific domain and the execution environment by automatically mapping high-level workflow descriptions onto distributed resources. It automatically locates the necessary input data and computational resources for workflow execution. Pegasus enables scientists to construct workflows in abstract terms without worrying about the details of the underlying execution environment or the particulars of the low-level specifications required by the middleware (Condor, Globus, or Amazon EC2). Pegasus WMS also bridges the current cyberinfrastructure by effectively coordinating multiple distributed resources. The input to Pegasus is a description of the abstract workflow in a YAML format.
Pegasus allows researchers to translate complex computational tasks into workflows that link and manage ensembles of dependent tasks and related data files. Pegasus automatically chains dependent tasks together so that a single scientist can complete complex computations that once required many different people. New users are encouraged to explore the Tutorial to become familiar with how to operate Pegasus for their own workflows in which users create and run a sample project to demonstrate Pegasus capabilities.
Pegasus has a number of features that contribute to its useability and effectiveness.
Portability / Reuse
User created workflows can easily be run in different environments without alteration. Pegasus currently runs workflows on top of Condor, Grid infrastructures such as Open Science Grid, Amazon EC2, Nimbus, and many campus clusters. The same workflow can run on a single system or across a heterogeneous set of resources.
Performance
The Pegasus mapper can reorder, group, and prioritize tasks in order to increase the overall workflow performance.
Scalability
Pegasus can easily scale both the size of the workflow and the resources that the workflow is distributed over. Pegasus runs workflows ranging from just a few computational tasks up to 1 million. The number of resources involved in executing a workflow can scale as needed without any impediments to performance.
Provenance
By default, all jobs in Pegasus are launched via the kickstart process that captures runtime provenance of the job and helps in debugging. The provenance data is collected in a database, and the data can be summarized with tools such as pegasus-statistics, pegasus-plots, or directly with SQL queries.
Data Management
Pegasus handles replica selection, data transfers, and output registrations in data catalogs. These tasks are added to a workflow as auxiliary jobs by the Pegasus planner.
Reliability
Jobs and data transfers are automatically retried in case of failures. Debugging tools such as pegasus-analyzer helps the user to debug the workflow in case of non-recoverable failures.
Error Recovery
When errors occur, Pegasus tries to recover when possible by retrying tasks, retrying the entire workflow, providing workflow-level checkpointing, re-mapping portions of the workflow, trying alternative data sources for staging data, and, when all else fails, by providing a rescue workflow containing a description of only the work that remains to be done. It cleans up storage as the workflow is executed so that data-intensive workflows have enough space to execute on a storage-constrained resource. Pegasus keeps track of what has been done (provenance) including the locations of data used and produced, and which software was used with which parameters.
Operating Environments
Pegasus workflows can be deployed across a variety of environments:
Local Execution
Pegasus can run a workflow on a single computer with Internet access. Running in a local environment is quicker to deploy as the user does not need to gain access to multiple resources in order to execute a workflow.
Condor Pools and Glideins
Condor is a specialized workload management system for compute-intensive jobs. Condor queues workflows, schedules, and monitors the execution of each workflow. Condor Pools and Glideins are tools for submitting and executing the Condor daemons on a Globus resource. As long as the daemons continue to run, the remote machine running them appears as part of your Condor pool. For a more complete description of Condor, see the Condor Project Pages
Grids
Pegasus WMS is entirely compatible with Grid computing. Grid computing relies on the concept of distributed computations. Pegasus apportions pieces of a workflow to run on distributed resources.
Clouds
Cloud computing uses a network as a means to connect a Pegasus end user to distributed resources that are based in the cloud.
1.1. What are scientific workflows?
A scientific workflow, or workflow, is an abstraction used by scientists to express an ensemble of complex, computational operations. In this context, an operation refers to a computational act such as retrieving data from remote storage services, executing applications, and transferring data products to designated storage sites.
Here we define key terminology within the context of Pegasus workflows which will be used throughout this chapter.
- job
A job (also referred to as a task) is the core entity in Pegasus in terms of execution. It encapsulates the executable that will be run (e.g. a Python script, an MPI application, a Java executable, a bash shell script, etc.), command line arguments that can be passed to the executable at runtime, required input files, and output files that will be produced.
- file
A file is the core entity in Pegasus in terms of data. Jobs (more specifically, the executable encapsulated by the job) will use as input zero or more files and will produce zero or more files.
- dependency
A dependency is a directed link between two jobs denoting a temporal or data dependency. A temporal dependency between two jobs means that the source job must complete before the destination job may run. A data dependency means that one or more of the output file(s) produced by the source job will be used as input in the destination job. For example, if “Job 1” writes a file which is then read by “Job 2”, then you have a data dependency from “Job 1” to “Job 2”.
- workflow
A workflow is the collected organization of jobs, files, and dependencies.
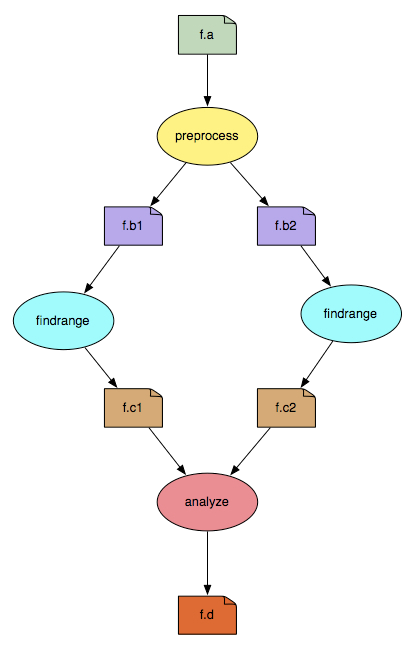
Figure 1: example workflow
To represent the computations imagined by scientists, workflows use a formal framework based on graph theory. A workflow is represented as a directed acyclic graph (DAG) whose nodes represent the jobs of the workflow (e.g., the tasks that need to be done) and the edges between those jobs represent dependencies (e.g., which jobs depend on which jobs). Figure 1 illustrates a simple workflow where ovals represent jobs and boxes represent files. An arrow from a file to a job is interpreted as “this job uses that file as input”. An arrow from a job to a file is interpreted as “this job produces that file”. For example, “preprocess -> f.b1 -> findrange” can be interpreted as “the job, preprocess, produces f.b1 as output, which is then used by findrange as input, and therefore a data dependency exists between preprocess and findrange”.
The execution of workflows, represented as DAGs, follows two main rules:
A job is considered finished when all its output files have been written.
A job cannot start before all its predecessors have finished their executions.
A workflow management system (WMS), such as Pegasus, is responsible for managing the execution of such workflows. It provides guarantees that jobs comprising the workflow will be executed in a sequence that is a valid topological ordering of the workflow. Furthermore, Pegasus provides a number of additional functionalities such as the handling of the movement of data products used/produced during the workflow execution, fault tolerance, and monitoring.
1.2. How to Convert Existing Applications Into a Workflow
The scientific community has long developed large applications, experiments, and analysis pipelines as workflows rather than monolithic entities because they are easier to manage and maintain. Furthermore, the DAG structure of the workflow exposes parallel regions within the application that can be taken advantage of using distributed and high performance computing resources. If you find that the codes you’ve developed need increasingly more resources to run, it can be advantageous to start adopting this “workflow” model. Then, a workflow management system like Pegasus can handle the execution of your codes at scale.
Converting an existing monolithic application/computational experiment into a workflow is fairly straightforward as most applications have an inherent DAG structure. Simply speaking, an application is just a set of functions executed in sequence, one after another. Consider the following Python script as a toy example of a monolithic application.
def preprocess(data):
# process the data
return data_processed1, data_processed2
def findrange(data):
# perform some computations
return data_range
def analyze(data1, data2):
# process (analyze) data1 and data2
return final_result
if __name__=="__main__":
data = [1,2,3,4,5,6]
data_processed1, data_processed2 = preprocess(data)
data_range1 = findrange(data_processed1)
data_range2 = findrange(data_processed2)
result = analyze(data_range1, data_range2)
print(result)
In this example, a Python function is similar to a job in Pegasus. Each function serves its own purpose and does some units of computation. Functions typically require some input and produce some output. That output is then consumed by another function, and so on and so forth. The major difference between a program and a job is that within the program, data objects may be passed between functions (shared address space) while in Pegasus, jobs are separate entities and cannot communicate between each other directly and thus must communicate using files.
Figure 1 illustrates what the above monolithic application would look like if it were to be translated into a workflow. The following steps outline what must be done to accomplish this translation:
Identify the functions or part of codes that are independent (i.e., functions that do not depend on any other functions);
Break the code into multiple scripts, each of them embedding one of the independent functions found in the previous step. In our example, we will have three scripts, one with the function “preprocess”, one with the function “findrange” and one with the function “analyze”;
If the function which was converted into a script has arguments, the script must be modified to read those arguments from a file(s).
If the function which was converted into a script returns data, the script must be modified to write that data to a file(s).
Use the Pegasus API, described in the following chapters, to link these independent scripts together and produce a Pegasus workflow.
1.2.1. Using Pegasus to Create the Workflow
Say that we’ve now broken apart the monolithic script from above into individual components. We would end up with a project directory that looks something like this:
project/
├── bin
│ ├── analyze.py
│ ├── findrange.py
│ └── preprocess.py
└── input_data
└── data.csv
Using the Python API provided by Pegasus, we can build and execute these codes as a workflow. The following snippet illustrates this:
#!/usr/bin/env python3
import logging
from pathlib import Path
from Pegasus.api import *
logging.basicConfig(level=logging.DEBUG)
# --- Specify Input Files ------------------------------------------------------
input_data = File("data.csv")
rc = ReplicaCatalog().add_replica(
site="local", lfn=input_data, pfn=Path(".").resolve() / "input_data/data.csv"
)
# --- Specify Executables ------------------------------------------------------
preprocess = Transformation(
name="preproces.py",
site="local",
pfn=Path(".").resolve() / "bin/preprocess.py",
is_stageable=True,
)
findrange = Transformation(
name="findrange.py",
site="local",
pfn=Path(".").resolve() / "bin/findrange.py",
is_stageable=True,
)
analyze = Transformation(
name="analyze.py",
site="local",
pfn=Path(".").resolve() / "bin/analyze.py",
is_stageable=True,
)
tc = TransformationCatalog().add_transformations(preprocess, findrange, analyze)
# --- Build Workflow -----------------------------------------------------------
wf = Workflow("analysis-workflow")
fb1 = File("f.b1")
fb2 = File("f.b2")
preprocess_job = (
Job(preprocess)
.add_args("arg1", "arg2")
.add_inputs(input_data)
.add_outputs(fb1, fb2)
)
fc1 = File("f.c1")
findrange_job1 = Job(findrange).add_inputs(fb1).add_outputs(fc1)
fc2 = File("f.c2")
findrange_job2 = Job(findrange).add_inputs(fb2).add_outputs(fc2)
result = File("result.csv")
analyze_job = Job(analyze).add_inputs(fc1, fc2).add_outputs(result)
wf.add_replica_catalog(rc)
wf.add_transformation_catalog(tc)
wf.add_jobs(preprocess_job, findrange_job1, findrange_job2, analyze_job)
From this script, we can see that all components in the
monolithic script have been accounted for. Using the
reference to the Workflow which was just created, it can then be executed with
wf.plan(submit=True). Continue to the Tutorial for a hands on
lesson in developing and running Pegasus workflows.
Note
We recommend visiting the Pegasus Workflow Repository or the Pegasus Application Showcase for examples of real-world workflows from a diverse set of scientific domains.