 Scientists at OSU use Pegasus for mass-spectrometry-based proteomics. Proteomics workflows have been executed on local clusters and cloud resources.
Scientists at OSU use Pegasus for mass-spectrometry-based proteomics. Proteomics workflows have been executed on local clusters and cloud resources.
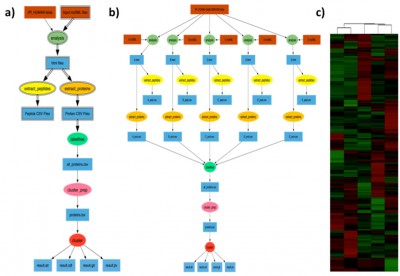
Example proteomic workflow: a) Pegasus workflow template. Square boxes with double lines represent file collections and the ellipses with double boundary represent parallel jobs. b) Implementation of workflow for clustering of five shotgun proteomic data sets. c) Hierarchical cluster analysis of the shotgun proteomic data.
Scientists: Michael Freitas, OSU