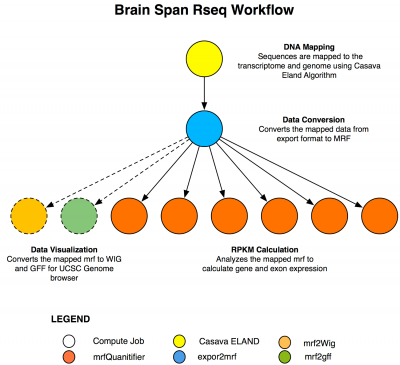
The Brain Span project seeks to find when and where in the brain a gene is expressed. This information holds clues to potential causes of disease. A recent study found that forms of a gene associated with schizophrenia are over-expressed in the fetal brain. To make such discoveries about what is abnormal, scientists first need to know what the normal patterns of gene expression are during development. To this end, the National Institute of Mental Health (NIMH), part of the National Institutes of Health (NIH), has funded the creation of TADHB. To map human brain “transcriptomes”, researchers identify the composition of intermediate products, called transcripts or messenger RNAs, which translate genes into proteins throughout development.
As part of this project we have enabled the geneticists to analyse over 225 human brain RNA sequences using two different mapping algorithms CASAVA ELAND and Perm.
As part of this project a Reusable Virtual Machine has been released with all the code and workflows to easily run the Workflow pipeline on their own RNA dataset. More information and VM download is availabe at http://genomics.isi.edu/rnaseq.
Publications: RseqFlow: Workflows for RNA-Seq data analysis Ying Wang, Gaurang Mehta, Rajiv Mayani, Jingxi Lu, Tade Souaiaia, Yangho Chen, Andrew Clark, Hee Jae Yoon, Lin Wan, Oleg V. Evgrafov, James A. Knowles, Ewa Deelman, and Ting Chen, Bioinformatics (2011) first published online July 27, 2011 doi:10.1093/bioinformatics/btr441