DIPA is ambitiously meant to be a pipedream for processing and analyzing diffusion weighted magnetic resonance imaging data. The processing involves various machine learning based signal distortion corrections, biophysical diffusion model estimation using non-linear optimization techniques, quality control, image registration using differential geometry both for transformation models and interpolation, region of interest extraction, connectivity estimation and statistical analysis using permutation testing and other advanced high-dimensional regression methods.
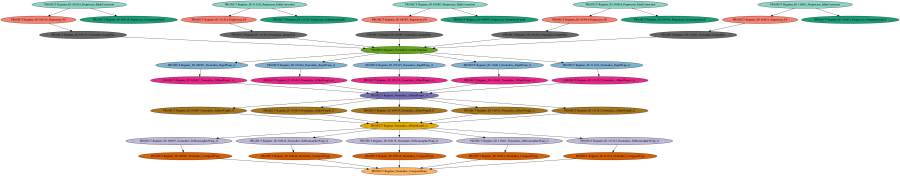
Typically, these types of processing pipelines and analysis on run almost every study at Waisman Center and ADRC (Alzheimer’s disease research center) which has diffusion MRI data. The number of scans can range from 100 to 500 and the pipelines are used to analyze publicly available data such as that from the human connectome project (HCP) which has 1000s of scans. Often these pipelines are run more than once per study, as subjects/scans are added.
GitHub Repository: https://github.com/pegasus-isi/dipa-workflow
Scientists: