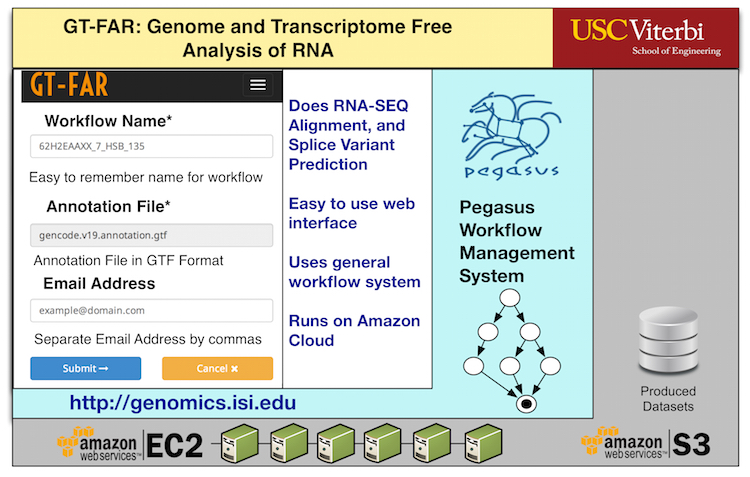
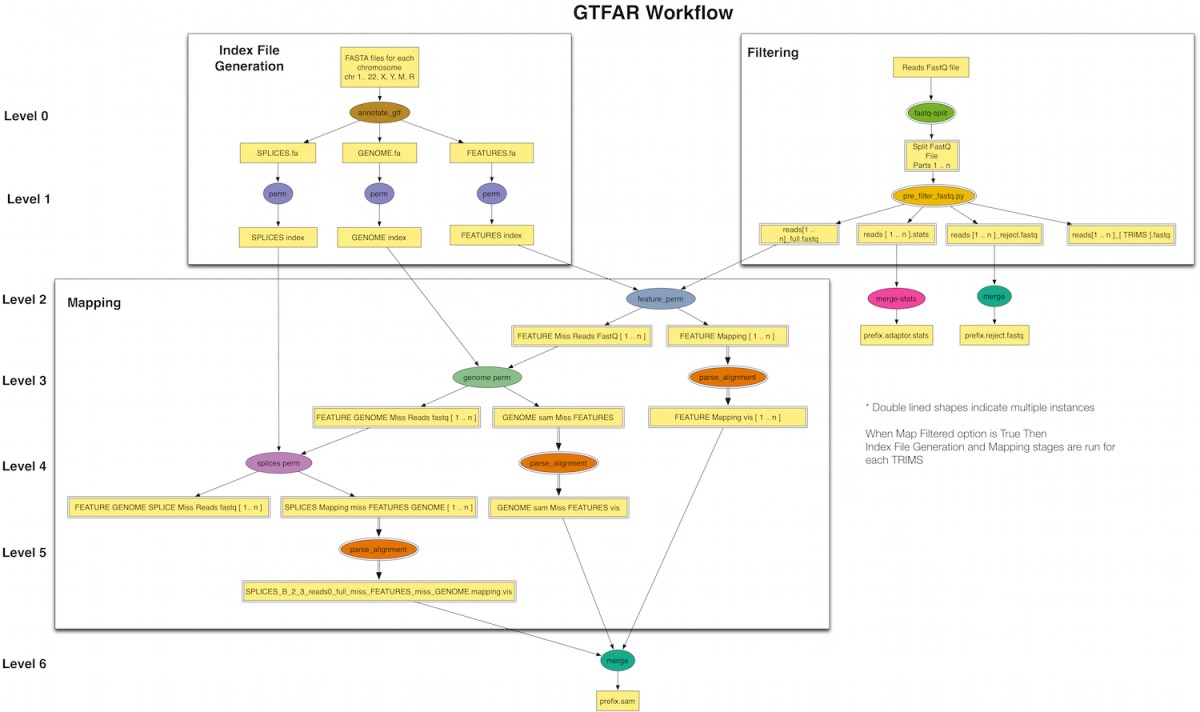
GT-FAR is a RNA seq pipeline that allows users to do Alignment, Quantification, Differential Expression, and Variant Calling. This pipeline has been modeled as a Pegasus workflow. Pegasus enables users to execute the pipeline on wide variety of execution environments ranging from local clusters, grids to computational clouds.
With funding support from iSeqTools , we have packaged this pipeline along with Pegasus as a cloud based solution. This Amazon based cloud solution, allows users to start a virtual machine on AmazonEC2, and upload their data-sets to perform GT-FAR analysis and makes outputs available in Amazon S3. The interface, allows scientists to run and monitor their analysis runs. It also generates error reports, that can be used to communicate back to developers if the analysis fail.
The web interface for the cloud based solution allows users to
- to upload custom datasets,
- start and monitor the progress of their GT-FAR analysis
- receive notifications when the analysis is done.
- make the output datasets available for download from S3.
Instructions for starting the Pegasus GT-FAR Amazon EC2 virtual image can be found here.
More information on pipeline: http://genomics.isi.edu/gtfar