 Plant scientists at University of Wisconsin Madison are using Pegasus to generate movies of plant root growth and analyze images collected via time-lapse photography. Another project samples forest locations to characterize the understory vegetation to determine how different plant species are distributed in the woods.
Plant scientists at University of Wisconsin Madison are using Pegasus to generate movies of plant root growth and analyze images collected via time-lapse photography. Another project samples forest locations to characterize the understory vegetation to determine how different plant species are distributed in the woods.
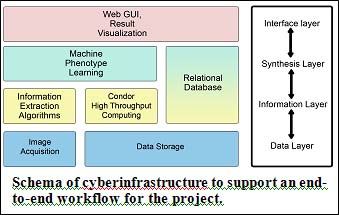
Edgar Spalding, a Professor of Botany and Faculty Affiliate of Biomedical Engineering is processing electronic images with computer algorithms can render some important aspects of plant development into a numerical format compatible with bioinformatics analysis. For example, the midline of an object, such as a root or a stem, contains much information about the size and shape of the object. Quantifying midline length and curvature distribution is one means of quantifying the size and shape of the object. The diagram shows a seedling root responding to gravity at three points in time. A midline is calculated. Curvature is calculated at each point along the midline, color-coded, and superimposed on the midline. Acquiring images every two minutes enables the biology to be quantitatively described with high spatiotemporal resolution. High throughput data acquisition coupled with high throughput computing makes genome-scale experiments feasible. A workflow (left) involving Pegasus WMS and UW Madison’s CHTC has processed hundreds of thousands of images and in some projects performed the statistical calculations required for genetic mapping, the first steps in identifying genes responsible for the quantified features. More about this project can be found at their website: http://www.botany.wisc.edu/spalding.htm