As the second OWM (old world monkey) sequenced (the first is Rhesus macaque), vervets, unlike the great apes who are mostly in near-extinction status, are widely available for biomedical research. (Rhesus is widely available in India but the export restriction imposed by the Indian government makes it less ideal for biomedical research). Given the genetic proximity to human compared to other model organisms such as mouse, vervet is a great model to study high-level cognitive traits, such as novelty-seeking, ADHD (Attention deficit hyperactivity disorder), etc. and some primate-specific diseases, such as HIV. A large pedigree of vervet monkeys housed in VRC (Vervet Research Colony) at North Carolina offers a great genetic resource to study these various traits in a controlled environment. Our focus right now is to sequence a large number of VRC monkeys with well-characterized and highly heritable phenotypes. The purpose is to find the genetic loci underlying these phenotypes.
On the other hand, the VRC was founded by 57 monkeys from two Caribbean islands, St. Kitts and Nevis, in late 1970s. St. Kits and Nevis have seen the population of vervet monkeys growing since they were brought to these island from west Africa through slave trade 300-400 years ago. So one natural question is to ask where their source is in Africa and how much genetic diversity has been lost. Furthermore, DNAs of wild vervet monkeys (5 different subspecies) from all over Africa have been collected to study the origin of vervet ancestry, the migratory pattern, and the genetic diversity of the entire vervet species. Insights from these population genetic studies serve as a foundation for future studies on the traits of wild vervet populations.
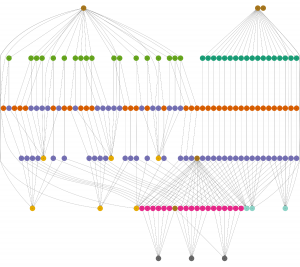
The workflow starts from next-generation sequence (NGS) data (100bp paired-end reads) from hundreds of Afican vervet monkeys (~120 currently, 3.5T in gzipped fastq file). It has a couple of sub-workflows. The alignment sub-workflow includes read filtering, read mapping, and duplication-marking. After this, a variant calling sub-workflow which involves several different current genotyping calling programs, i.e. GATK, SAMtools, Triocaller, is applied in order to take intersection of them. The example shown in the picture is this variant calling workflow running on a small input. After this step, Pegasus was further used to construct workflows for variant filtering, variant comparison, calculating population genetics measures, etc.
http://crocea.mednet.ucla.edu/
Scientists:
Yu S. Huang et al. (Center for Neurobehavioral Genetics, UCLA)