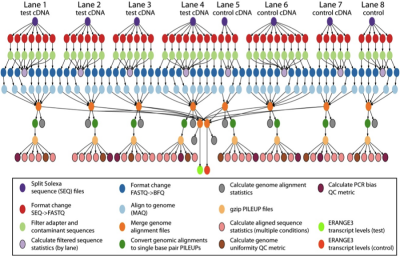
 This application splits sequence files into multiple parts and converts them to the appropriate file format. Then it filters out noisy and contaminating sequences to maps them to their genomic locations. From the individual mapping steps, it merges them into a single global map and uses sequence maps to calculate the sequence density at each position in the genome. This application has a footprint of 6GB of data.
This application splits sequence files into multiple parts and converts them to the appropriate file format. Then it filters out noisy and contaminating sequences to maps them to their genomic locations. From the individual mapping steps, it merges them into a single global map and uses sequence maps to calculate the sequence density at each position in the genome. This application has a footprint of 6GB of data.
Scientists: Ben Berman et al. (USC)