With the advances in next generation sequencing (NGS) technology and significant reduction in sequencing costs it is now possible to sequence large sets of crop germplasm and generate whole genome scale structural variations and genotypic data. In depth informatics analysis of the genotypic data can provide better understanding of the links with the observed phenotypic changes. This approach can be used to further understand and study different traits for the improvement of crops by design. The NGS resequencing data represents a rich source of information and can lead to significant discoveries when it comes to mining genotypic data for phenotypic inferences.
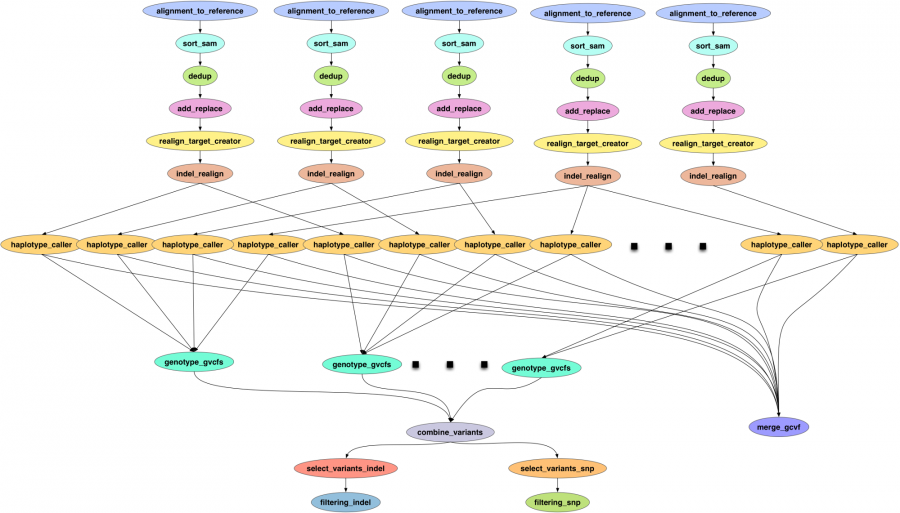
The NGS reseqeuncing data is hosted in the CyVerse Data Store, which is based on iRODS. The Data Store is mainly hosted at the CyVerse center at the University of Arizona. An advantage to this setup has been that TACC is a CyVerse site and has dedicated storage servers for the CyVerse Data Store. Data can be replicated from Arizona to the servers at TACC, which allows low latency data access to the inputs and output data store when running the workflows on the TACC Wrangler resource. The Pegasus workflow system is used to define and control the required computational tasks. This includes the user defined tasks, such as BWA, Picard, and GATK, as well as Pegasus added tasks such as data staging between the CyVerse Data Store and the Wrangler flash based scratch filesystem. Pegasus also adds data cleanup tasks to maintain and minimize the workflow footprint on the scratch filesystem as the workflow progresses.
Following this GATK workflow, downstream analysis including copy number variations (CNV) analysis using cn.MOPS, SNPs annotations using SnpEff and SnpSift, linkage disequilibrium for haplotype identification using LDExplorer and hierarchical tree generation using SNPViz are also finished. All data will be applied for GWAS phenotype-genotype study, comprehensive traits analysis and soybean breeding program. Data and results can be accessed through web-based Soybean Knowledge Base (SoyKB) at http://soykb.org, hosted at CyVerse. The integrated workflow can be easily customized to other species with large-scale NGS data as well.
The Pegasus workflow can be found on GitHub: https://github.com/pegasus-isi/Soybean-Workflow