Time: 2pm-5pm
Date: Tuesday, March 22nd, 2016
Location: Aresty Auditorium at Health Sciences Campus
Instructor: The Pegasus team from Viterbi’s Information Sciences Institute
Registration: https://uscits.az1.qualtrics.com/SE/?SID=SV_8p7zWppguSYbXKJ
Course Material: https://pegasus.isi.edu/tutorial/usc/
The Pegasus Team is hosting a half day workshop on March 22nd, 2016 at the Health Science Campus. This workshop includes a hands-on component that requires an active HPC account. If you don’t have USC HPC account and want to attend the workshop, HPC team can now offer temporary HPC accounts for workshop attendees. To be eligible, you must have a USC NetID, and must register via the Registration link below. This is a great way to check out HPC and learn about Workflows if you do not have an HPC account.
Scientific Workflows via The Pegasus Workflow Management System on the HPC Cluster
Workflows are a key technology for enabling complex scientific applications. They capture the interdependencies between processing steps in data analysis and simulation pipelines, as well as the mechanisms to execute those steps reliably and efficiently in a distributed computing environment. They also enable scientists to capture complex processes to promote sharing and reuse, and provide provenance information necessary for the verification of scientific results and scientific reproducibility.
In this workshop, we will focus on how to model scientific analysis as a workflow that can be executed on the USC HPC cluster using Pegasus WMS (http://pegasus.isi.edu). Pegasus allows users to design workflows at a high-level of abstraction, that is independent of the resources available to execute them and the location of data and executables. It compiles these abstract workflows to executable workflows that can be deployed onto distributed resources such local campus clusters, computational clouds and grids such as XSEDE and Open Science Grid. During the compilation process, Pegasus WMS does data discovery, whereby it determines the locations of input data files and executables. Data transfer tasks are added to the executable workflow that are responsible for staging in the input files to the cluster, and the generated output files back to a user specified location. In addition to the data transfers tasks, data cleanup (cleanup data that is no longer required) and data registration tasks (catalog the output files) are be added to the pipeline.
Through hands-on exercises, we will cover issues of workflow composition, how to design a workflow in a portable way, workflow execution and how to run the workflow efficiently and reliably on the USC HPC cluster. An important component of the tutorial will be how to monitor, debug and analyze workflows using Pegasus-provided tools. The workshop will also cover how to execute MPI application codes as part of a workflow.
This workshop is intended for both new and existing HPC users. It is highly recommended that you take the Introduction to Linux/Unix workshop if you haven’t worked in the Linux environment before. The participants will be expected to bring in their own laptops with the following software installed: SSH client, Web Browser, PDF reader. If you have any questions about either of these workshops, please send email to hpc@usc.edu and erinshaw@usc.edu. We look forward to seeing you there!
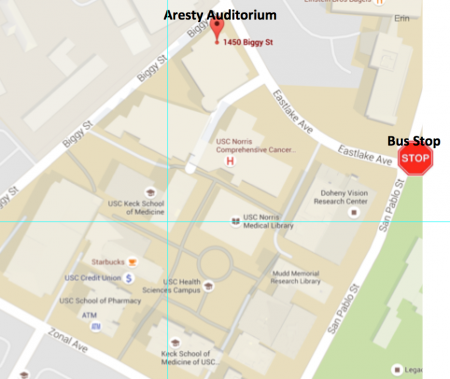
How to get there
Transportation (see map below): If you will be taking the Inter-Campus Shuttle to HSC, we recommend the 1pm shuttle that leaves from JEP House and 34th St./McClintock and drops off at Eastlake & San Pablo (because of construction; so this may change). Aresty Auditorium is accessible through Norris Comprehensive Cancer Center. It is street level and there is also a street entrance. (However, on the morning of our previous workshop the doors were locked, maybe because it was before 8:30am. Just FYI. We will keep an eye out for those entering that way if they are not open)